getwd()Week 1 Lab: Introduction to R and Tableau
Learning Objectives
This lab contributes to Course Objectives 2, 3, & 4
Open R/Rstudio and ensure it is functioning properly
Get oriented with RStudio
Navigate directories from within R
Conduct basic calculations in R
Read in an external data set in R
Load a package in R
Create your Google site and a project page
Apply for Tableau academic license
Download Tableau desktop (if using personal computers) and ensure working correctly
Register for Tableau public
Download Lab Handout Here
Introduction to R
Welcome to the world of R programming! R is a powerful and widely used programming language in the field of data analysis and statistical computing. It is known for its vast collection of libraries and packages that allow users to easily perform complex data manipulation and analysis tasks.
Before lab 1, if you are using your personal computer, you will need to install R and R Studio on your computer.
Step 1: Installing R: You can download the latest version of R for free from the official website https://cran.r-project.org/.
Step 2: Installing RStudio: Once the installation is complete, you will also need to install RStudio, a popular integrated development environment (IDE) for R. You can download RStudio from the official website https://rstudio.com/.
RStudio provides a user-friendly interface for working with R and offers a variety of features to make working with R easier and more efficient. Open up RStudio and you should see something that looks like this:
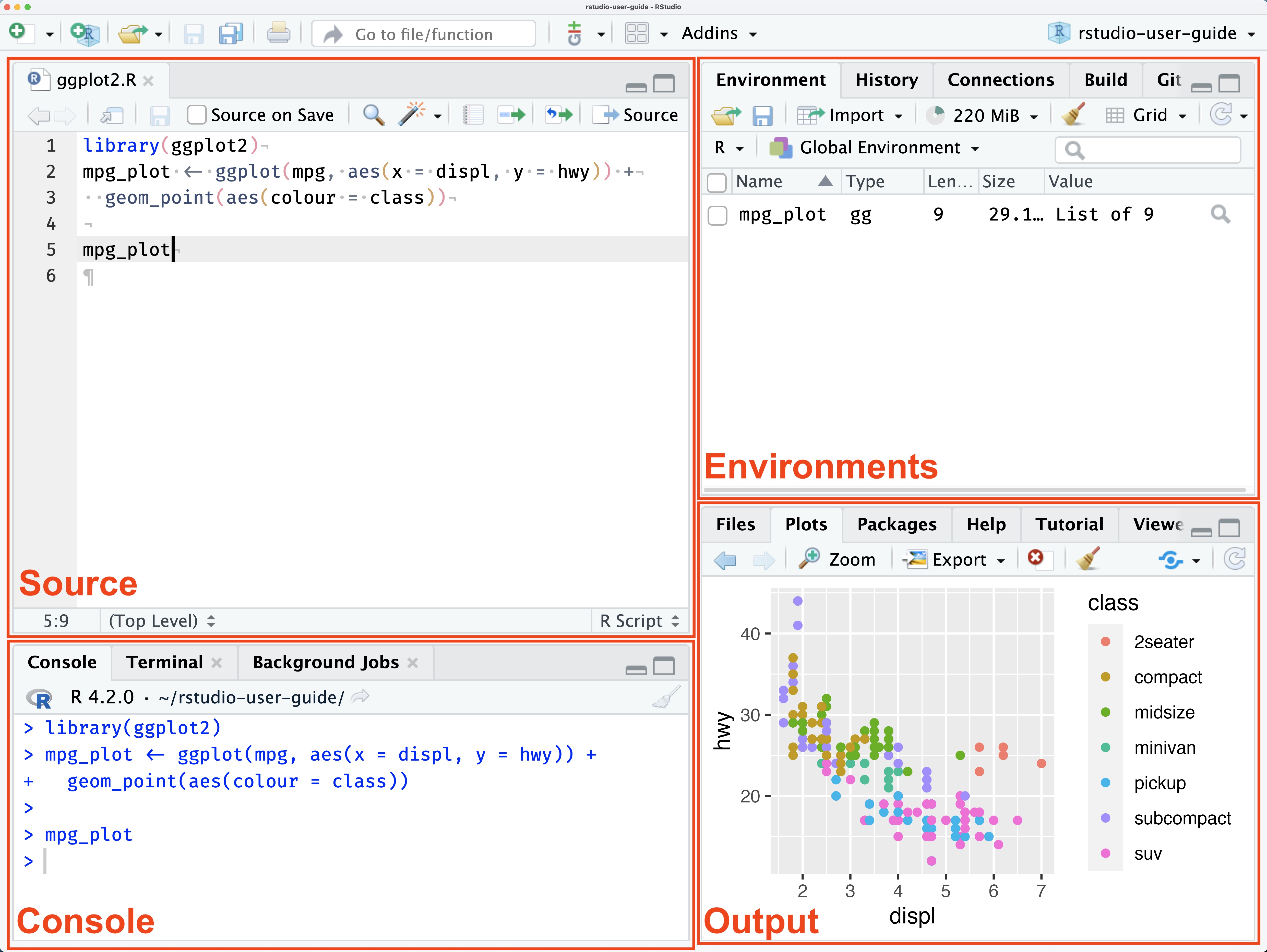
The RStudio interface is divided into several panels:
The Console panel is where you can enter and run R commands.
The Source panel is where you can open and edit R scripts, which are files that contain R code.
The Environment and History panels display information about the current environment and command history.
The Output panel includes several tabs (Files, Plots, Packages, and Help) that provide access to various resources and tools.
Now that you have both R and RStudio installed and running, let’s dive into some basic concepts of the R programming language.
Opening a new R script
Because R is a command-line application, we can write a script (i.e., text file) with a sequence of commands. There are many benefits to scripting:
- You can save a record of the commands if you need to reproduce the analysis.
- You can share it with others so they can reproduce it.
- You can easily change a part of your script to see what happens.
Locating your working directory
The working directory is the folder on your computer where R looks for files to read and where it saves any files you create. It’s like the “home base” for your R session. Knowing your working directory is important because:
- If you want to read a data file (like a
.csvfile), R will look for it in the working directory unless you provide a full path. - When you save files, such as plots or datasets, R will save them in the working directory by default.
Think of it as the default folder R uses to organize your work during a session.
You can check your current working directory by using the getwd() function:
This will display the path of the current working directory in the console.
To set your working directory to a specific folder, use the setwd() function and provide the path to the folder. For example:
setwd("C:/Users/YourName/Documents/MyRProjects")After running this command, R will use the specified folder as the working directory for the rest of the session.
Tips for managing your working directory
In RStudio, you can also set the working directory interactively by going to Session > Set Working Directory > Choose Directory… and selecting the folder.
You can see the contents of your working directory in the Files panel in RStudio.
For reproducibility, it’s a good idea to include the
setwd()command in your R script so that others can run your code without confusion about file locations.
Think of the working directory as a specific “drawer” in your desk where you store tools. When you start working, you know where to find your materials and where to put things back.
R Basics
R is an interpreted language, which means that you can enter commands directly into the console and see the results immediately. You can start the R console by opening RStudio and clicking on the “Console” tab.
Defining variables
To assign a value to a variable in R, you can use the assignment operator <- or = like this:
x <- 5
x = 5 #Does the same thingHere, we have created a variable called x and assigned it the value of 5. Once you assign a variable a value, you can see it in your environment panel. You can use the print() function to display the value of a variable:
print(x)[1] 5When you have multiple pieces of code in a single code chunk (or even a single piece of code), it is recommended that you use code comments to explain what your code is doing. Since R treats everything inside a code chunk as code, if you want to write some text comments inside a code chunk, you can tell R that your text is not code by placing a # symbol at the beginning of your text like this:
# compute 4 times 5
4 * 5[1] 20R will ignore anything that follows a # symbol. So in the above code chunk, R will ignore the first line with the code comment “compute 4 times 5”, and then it will compute the R code on the next line, 4 * 5.
Code comments are really helpful for explaining what your code is doing. I usually reserve the text outside code chunks for more general discussion of my data, analysis, and results and I reserve code comments inside code chunks for explaining my code itself. Since I tend to forget the reasons behind certain decisions I made in my code, adding explanations in code comments helps me remember my motivations and intentions days, months, or even years later.
Doing math with R
You can also perform basic arithmetic operations in R using the standard operators (+, -, *, /). Here are some helpful math symbols in R:
- Parentheses:
(,) - Exponents:
^or** - Multiply:
* - Divide:
/ - Add:
+ - Subtract:
-
Some basic mathematical computations you can compute in R:
y <- 3
z <- x + y
print(z)[1] 8First, we assign the variable y the value 3. Then, we assign z the value of x (assigned 5 earlier) plus y (just assigned 3), which sum to 8.
Try assigning the variables a and b different values and dividing them.
Creating vectors
R also has a wide range of data types, including: numerical, character, and logical. Variables can be different datatypes. Collections of variables are called a vector - a one-dimensional array of data. You can create a vector using the “c()” function, which stands for concatenate. A vector may only contain one type of data.
numbers <- c(1, 2, 3, 4, 5)
characters <- c("a", "b", "c")
logical <- c(TRUE, FALSE, TRUE)Indexing
A single entry in a vector is called an element. You can access elements of a vector using the square bracket notation:
print(numbers[1])[1] 1Try accessing a different element of the vector numbers or one of the other vectors.
You can also access series of elements in the vector. To access a continuous range of values use the colon :
print(characters[2:3])[1] "b" "c"You can even use a vector of index values to call discontinuous subsets of the vector.
print(characters[c(1,3)])[1] "a" "c"Depending on the datatype, there are functions that operate on the vector. For example, you can sum the elements of the numeric vector numbers.
print(sum(numbers))[1] 15Can you sum the numbers of the vector characters? How about logical?
You can also add, subtract, multiply, and divide all of the numbers in a vector
print(numbers+5)[1] 6 7 8 9 10Or add, subtract, multiply, and divide numbers of equal-length vectors
print(numbers + numbers*2)[1] 3 6 9 12 15Can you figure out how to calculate the product of the vector numbers?
Building data frames
Using R like a fancy calculator is intended to help you understand how R works. R is generally used to analyze data sets. Data sets are collections of equal-length vectors in a table or what R calls a data.frame. Here is a simple example:
print(data.frame(a=c(1:3),
b=c(4:6),
d=c(7:9))) a b d
1 1 4 7
2 2 5 8
3 3 6 9Where a,b,d are the column or variable names and the 1,2,3 on the far left are row numbers.
You can build a data.frame using the vectors above and assign the object a name.
df <- data.frame(numbers=numbers[1:3],
characters,
logical)
print(df) numbers characters logical
1 1 a TRUE
2 2 b FALSE
3 3 c TRUEWhy did I subset the vector numbers?
Reading data
Creating data.frames by hand is tedious. Fortunately, R has some utilities to read data stored in certain files types. Before we explore R’s ability to read data, we must understand how R interacts with the computer’s file system.
At any point in time, you are in an active directory (similar to windows explorer or macos finder). You can ask R to tell you the current working directory (where R is looking at the moment).
getwd()[1] "C:/Users/lachenar/OneDrive - Colostate/Documents/GitProjectsWithR/csu-arec-330.github.io/materials/unit_00/week_01"The response to this command will depend on the directory structure on your machine.
You can navigate your directory structure using the command setwd(). When you have multiple projects with multiple files on your machine, it will be important that you understand R’s working directory and how to change it. The syntax will depend on whether you are on a windows or mac. First, locate the data file that you downloaded for this exercise. You will treat this like the project directory and instruct R to navigate your directory structure by typing the path to the directory in the setwd("directory/subdirectory1") command. The path needs to be in quotes to tell R that you are inputing characters and not variables.
This video describes a computer’s file system and organization: https://www.youtube.com/watch?v=hUW5MEKDtMM
To read data into R, we will use another function built into R. We will be reading a file type called comma-separated values denoted by the file extension .csv. The commas separate the variables, so R knows where a new variable starts. We will read the file into a data.frame called super_sales. You can download the file here.
super_sales <- read.csv("supermarket_sales.csv")You can give objects (i.e., variables, vectors, and data frames) almost any name as long as it does not start with a number and it cannot contain a space. However, you want to choose descriptive names so that your future self and others will find it easier to read. I prefer what is called snake_case where words are separated by an underscore and all lower case. There are other options and it is a matter of opinion. I suggest choosing one and sticking with it.
# Lab Script for Week 1: Introduction to R
# Set and check the working directory
getwd() # Displays the current working directory
setwd("C:/Users/YourName/Documents/MyRProjects") # Sets the working directory
# Assign values to variables
x <- 5 # Assign 5 to x using the preferred assignment operator
x = 5 # Does the same thing but using = for assignment
# Print the value of x
print(x)
# Perform basic arithmetic
# Compute 4 times 5
4 * 5
y <- 3 # Assign 3 to y
z <- x + y # Add x and y, assign the result to z
print(z) # Print the value of z
# Divide two variables
a <- 30 # Assign 30 to a
b <- 10 # Assign 10 to b
a / b # Divide a by b
# Create vectors of different data types
numbers <- c(1, 2, 3, 4, 5) # Numeric vector
characters <- c("a", "b", "c") # Character vector
logical <- c(TRUE, FALSE, TRUE) # Logical vector
# Access elements of a vector
print(numbers[1]) # Print the first element of numbers
print(numbers[3]) # Print the third element of numbers
print(characters[2:3]) # Print the second and third elements of characters
print(characters[c(1,3)]) # Print the first and third elements of characters
# Can you sum the numbers of the vector characters? How about logical?
# Attempting to sum the characters vector will result in an error because it contains non-numeric data.
# Uncomment the line below to see the error.
# sum(characters)
# Sum the logical vector. TRUE is treated as 1, and FALSE is treated as 0.
# print(sum(logical))
# Perform vectorized operations
print(numbers+5) # Add 5 to each element of numbers
print(numbers + numbers*2) # Multiply each element of numbers by 2 and add it to the original numbers
# Can you figure out how to calculate the product of the vector numbers?
# Use the prod() function to calculate the product of all elements in the vector
print(prod(numbers))
# Create and display a data frame
print(data.frame(a=c(1:3),
b=c(4:6),
d=c(7:9)))
# Create a data frame using existing vectors
df <- data.frame(numbers=numbers[1:3],
characters,
logical)
print(df) # Print the data frame
# Read in an external data set
super_sales <- read.csv("https://csu-arec-330.github.io/materials/unit_00/inputs/supermarket_sales.csv")
head(super_sales) # Display the first few rows of the data set
# Install and load a package
# Run the following command only once to install the package
# install.packages("dplyr")
library(dplyr) # Load the dplyr packageRunning scripts and saving output
In this class, you’ll often write scripts, which are text files containing a series of R commands (like a recipe for your analysis). Once your script is complete, you may want R to execute all the commands at once. The command to run a script in R is source().
You can run source() directly in the Console, or click the Source button in the upper-right corner of the Source panel in RStudio. For example, suppose you have a script named lab_01_example.R saved in your current working directory. You can run it by typing the following in the Console:
source("lab_01_example.R", echo = TRUE)echo = TRUE tells R to print each command and its output in the Console as the script runs, making it easier to follow what’s happening.
Creating a log file with sink()
For assignments in this class, you’ll need to demonstrate that your scripts run without errors. A helpful way to do this is to generate a log file that captures both the commands from your script and their output. You can create a log file using source() together with another function called sink().
Here’s how to generate a log file for a script named lab_01_example.R:
- Open a new script in RStudio.
- Type the following commands (lines) into the new script or directly into the Console:
sink(file = "lab_01_example.log")
source("lab_01_example.R",echo = TRUE)
sink()How it works:
sink(file = "lab_01_example.log"): This starts redirecting all output from R into a file namedlab_01_example.log.source("lab_01_example.R", echo = TRUE): Runs the script and ensures both the commands and their results are included in the log file.sink(): Closes the connection to the log file, returning R to normal behavior (printing output to the Console).
Once the script runs, you can open the log file (lab_01_example.log) to review everything that was executed and ensure your code ran without errors.
Watch the demo
Here is a video that walks you through how to generate a log file using sink() and source() in RStudio. You can also access the video through this link.
Packages or Libraries
R has a vast collection of libraries or packages that provide a wide range of functions for data manipulation and analysis. Since R is open source, anyone can write libraries and share them with the world.1 You can install and load a package using the “install.packages()” and “library()” functions:
#Run only once
#install.packages("dplyr")
library(dplyr)Warning: package 'dplyr' was built under R version 4.4.2
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionHere, we have installed and loaded the dplyr package, which provides a wide range of functions for data manipulation. Note that you only need to install packages once, but you need to load them using the library() command each session. If you are working on the server, the packages you need will be installed; you just need to load them.
Documentation
Documentation is critical to understanding how functions and packages work in R. There is a help tab in the lower right panel in RStudio. You can also place your cursor in the function name in your code and press F1 to bring up the help for that function. R documentation always provides a brief description, information about arguments (or inputs), information about outputs, and some examples of how you would use it. Learning to read the documentation is critical for learning new packages.
Here are some additional resources to help you get started with R:
Introduction and RStudio setup: https://youtu.be/dFSPmjSynCs
data.frames: https://youtu.be/ULjXUW5yeDM
packages and libraries: https://youtu.be/l5bmDv98zX4
Google Site
Google sites is an easy-to-use website editing and hosting service. You will create a website as a portfolio for your work in the course. You will complete your assignments by building webpages in your website. We encourage you to develop this website to showcase your work and market yourself to future employers.
Tableau Setup
We will be using two Tableau products in this course (although there are many more - see handout): Tableau Desktop and Tableau Public. Tableau desktop typically requires a paid subscription, but as students (and teachers) we get it for free.
Visit Tableau for students and apply for your 1-year academic license.
Visit Tableau Desktop and download Tableau Desktop using the 14-day free trial. You can enter your product license key once you receive an email with your academic license.
Visit Tableau Public and create an account. You will use this account to host your data visualizations, which you can then embed on your google site. You will be required to use this to upload homework assignments.
If you are ever feeling lost in this class, I recommend checking out some of Tableau’s free eLearning resources. We will use some videos from these trainings throughout the course and I am happy to recommend specific resources to fit your needs.
This Week’s Assignment
Get started on the problem set here: https://csu-arec-330.github.io/materials/unit_00/week_01/ps1.html